![\begin{figure}
\begin{center}
\scalebox {0.4}{\includegraphics*[02cm,18cm][28cm,28cm]{naiveb.eps}}\end{center}\end{figure}](img8.gif) |
Bayesian networks is a statistical method for Data Mining, a statistical method for discovering valid, novel and potentially useful patterns in data. A description of Probabilistic networks and machine learning techniques is given in [15].
Bayesian networks are used to represent essential information in databases in a network structure. The network consists of edges and vertices, where the vertices are events and the edges relations between events. A simple Bayesian network is illustrated in Fig 3.2, where symptoms are dependent on a disease, and a disease is dependent on age, work and work environment. Introductions to Bayesian networks can be found in [13], [25], [17], [14] and [18].
Bayesian networks are easy to interpret for humans, and are able to store causal relationships , that is, relations between causes and effects. The networks can be used to represent domain knowledge, and it is possible to control inference and produce explanations on a network.
A simple usage of Bayesian networks is denoted naive Bayesian classification [15]. These networks consist only of one parent and several child nodes as in Fig 3.3. Classification is done by considering the parent node to be a hidden variable (H in the figure) stating which class (child node) each object in the database should belong to. An existing system using naive Bayesian classification is AutoClass [29].
The theoretical foundation for Bayesian networks is Bayes rule, which states:
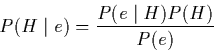
Where H is a hypothesis, and e an event. ![]() is the posterior
probability, and P(H) is the prior probability. A proof of Bayes rule is
given in appendix A.
is the posterior
probability, and P(H) is the prior probability. A proof of Bayes rule is
given in appendix A.
To give a formal definition of Bayesian networks, we introduce some terminology which is taken from [25]:
If a subset of Z nodes in a graph G intercepts all paths between the nodes
X and Y (written ![]() ), then this corresponds
to conditional independence between X and Y given Z:
), then this corresponds
to conditional independence between X and Y given Z:
![]()
conversely:
![]()
with respect to some dependency model M.
A Directed, Acyclic Graph (DAG) D is said to be a I-map of a dependency model M if for every three disjoint sets of vertices, X, Y and Z we have:
![]()
A DAG is a minimal I-map of M if none of its arrows can be deleted without destroying its I-mapness.
Given a probability distribution P
on a set of variables U, a DAG ![]() is called a Bayesian
Network of P iff
is called a Bayesian
Network of P iff![]() D is a minimal I-map of P.
D is a minimal I-map of P.
A Bayesian network is shown in Fig 3.3, representing the probability distribution P:
![]()